Drug Discovery, Population Genetics, Vaccine Design, and Computational Genomics.
![]()
Computational Biology
We use computational methods to analyze and study large segments of data on biological systems. These may include genetic sequences, protein samples, cell populations, and the like.
Here at the Ghansah Lab our focus areas under computational biology research include drug discovery, population genetics, vaccine design and computational genomics. These focus areas necessitate the application of computational methods which include but are not limited to mathematical modeling and simulation, machine learning, and artificial intelligence.

The Ghansah Lab team employs computational methods such as virtual screening and molecular docking
Vaccine Discovery
Project Title: In-silico design of vaccine candidates against Plasmodium falciparum
targeting the circumsporozoite protein
The malaria burden in endemic regions especially in sub-Saharan Africa still remains relatively high. One main tool lacking in the arsenal is a broadly effective vaccine against the most lethal causative parasite of malaria, Plasmodium falciparum. The circumsporozoite protein (CSP) has been one of the most promising target for vaccine design as the currently the most advanced malaria vaccine, RTS,S/AS01 was designed from the CSP antigen. The efficacy of RTS,S/AS01 vaccine was about 39%, which falls short of World Health Organisation (WHO) target of at least 75% vaccine efficacy. Using the CSP as target, we predicted immunogenic epitopes using Plasmodium falciparum 3D7 strain haplotypes in three populations in Ghana. This has uncovered some immunogenic leads as vaccine candidates that had over 85 percent population coverage for the three populations studied. Cape Coast, Kintampo and Navrongo. Moving forward, we want to explore these lead immunogenic epitopes in wet lab experimental designs for the development of broadly efficacious vaccines against malaria.
Drug Discovery
The team employs computational methods such as virtual screening and molecular docking to analyze 3D crystallographic structures of proteins of the Plasmodium falciparum parasite and predict or identify useful compounds that could inhibit protein functions or activate pathways that could mitigate the development of valuable proteins to the parasite.
We undertake Population Genetics research of Plasmodium falciparum where we study the genomic signatures of genes under natural selection. We study drug resistant genes and how these genes are different between different parasite populations. This helps to track the spread of these drug resistant genes in the understudy populations.
With a computational vaccine design approach, we predict antigenic regions (epitopes) from known vaccine target genes (antigens) of Plasmodium falciparum. The immunogenicity of these epitopes are evaluated through molecular docking and dynamics simulations with known human leukocytes antigens (HLAs) from the understudy population.
Computational drug discovery projects
The milestone to malaria elimination has been disrupted by drug resistant strains. This promotes the urgent need for novel antimalarial drug discovery and the uncovering of other crucial resistance mechanisms.
In-silico identification of natural product compounds to circumvent the activities of plasmodium Kelch 13 mutations in africa.
Recent research indicates that artemisinin resistance is conferred by mutations in the Plasmodium falciparum kelch13 (Pfk13) by inhibiting hemoglobin endocytosis. By analyzing these mutations to determine haplotypes in the African population, performing molecular interaction and dynamics studies, we could predict novel therapeutics with the potential to circumvent the activities of these mutations.
Leveraging computational approaches in drug discovery and resistance in Malaria
This research focuses on leveraging biomolecules/vital proteins in the Plasmodium genome using structural bioinformatics methods, ligand chemistry and dynamics (structure-activity relationship) to discover new compounds active against the parasite. We are additionally working on implementing pipelines for unveiling drug resistance mechanisms in parasite genes.
Computational Biology Articles
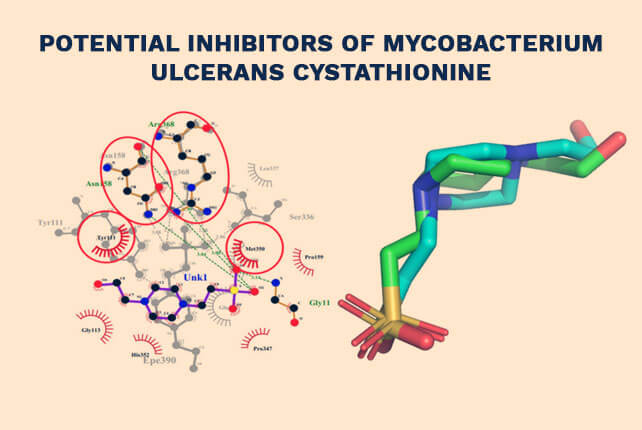
Potential Inhibitors of Mycobacterium ulcerans Cystathionine
Pharmacophore-Guided Identification of Natural Products as Potential Inhibitors of Mycobacterium ulcerans Cystathionine γ-Synthase MetB – 2021
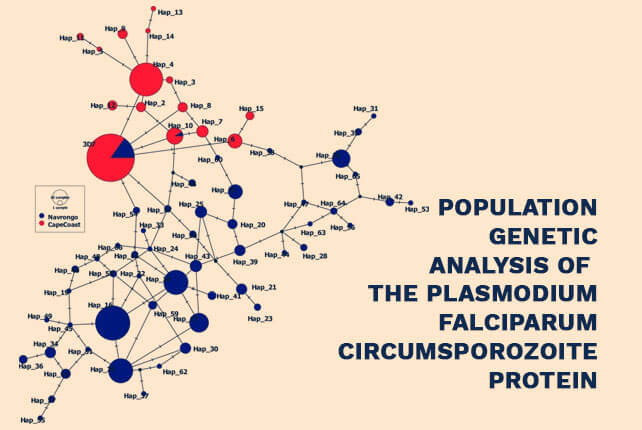
Population genetic analysis of the Plasmodium falciparum circumsporozoite protein
Population genetic analysis of the Plasmodium falciparum circumsporozoite protein in two distinct ecological regions in Ghana – 2020
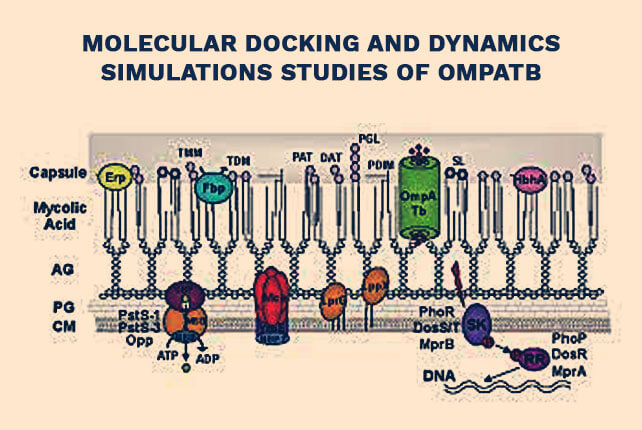
Molecular docking and dynamics simulations studies of OmpATb
Molecular docking and dynamics simulations studies of OmpATb identifies four potential novel natural product-derived anti-Mycobacterium tuberculosis compounds – 2020
COMPUTATIONAL RESEARCH PROJECTS
In-silico design of vaccine candidates against Plasmodium falciparum targeting the circumsporozoite protein
The malaria burden in endemic regions especially in sub-Saharan Africa still remains relatively high. One main tool lacking in the arsenal is a broadly effective vaccine against the most lethal causative parasite of malaria, Plasmodium falciparum.
The circumsporozoite protein (CSP) has been one of the most promising target for vaccine design as the currently the most advanced malaria vaccine, RTS,S/AS01 was designed from the CSP antigen. The efficacy of RTS,S/AS01 vaccine was about 39%, which falls short of World Health Organisation (WHO) target of at least 75% vaccine efficacy.
Using the CSP as target, we predicted immunogenic epitopes using Plasmodium falciparum 3D7 strain haplotypes in three
populations in Ghana. This has uncovered some immunogenic leads as vaccine candidates that had over 85 percent population coverage for the three populations studied.
Cape Coast, Kintampo and Navrongo.
Moving forward, we want to explore these lead immunogenic epitopes in wet lab experimental designs for the development of broadly efficacious vaccines against malaria.

Founders: Pine BioTech Team
Leverages big data in biomedical research, biotechnology and agriculture to help more people live happier and healthier lives.
Collaborator : The Bailey Lab from Brown University
The Bailey Lab is Advancing infectious disease research through genomics and genetics. Located in beautiful downtown Providence, Rhode Island; we are an incredible collaborative community working closely with investigators all over the world.
Recent News
Anita Ghansah Lab Presentation – Malaria Surveillance
https://youtu.be/TmfiephN7WY
Workshop on Varcode
Today we begin our varcode training with Dr. Anita Ghanasah and Dr. Kathryn Tiedje. Dr Kathryn Tiedje is a Research Fellow working in the laboratory of Prof. Karen Day based in the Peter Doherty and Bio21 Institutes at The University of Melbourne, AU. Kathryn is an...
Meeting with NMCP
The National Malaria Control Programme (NMCP) in collaboration with the Noguchi Memorial Institute for Medical Research (NMIMR) have worked together for many years to control Malaria in Ghana. Earlier this year, the NMCP gave the Ghansahlab the opportunity to...